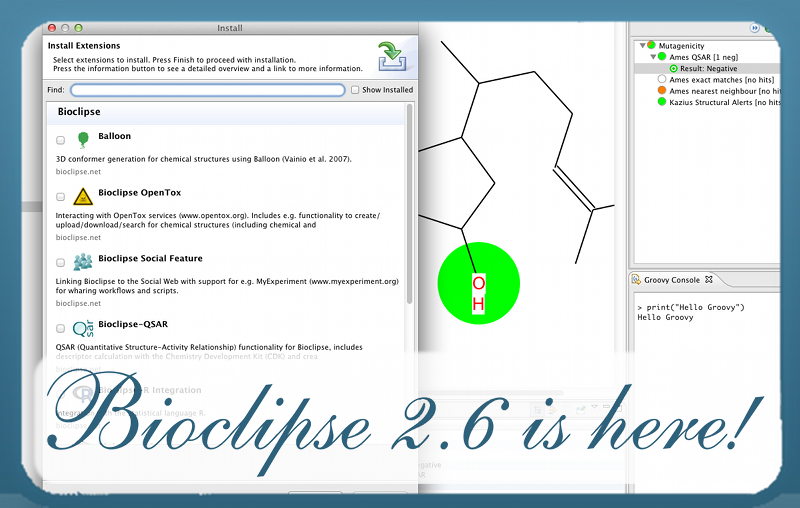
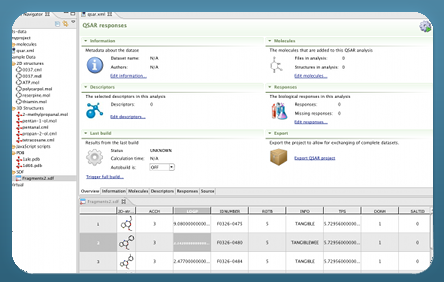
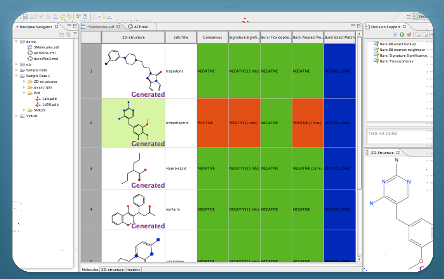
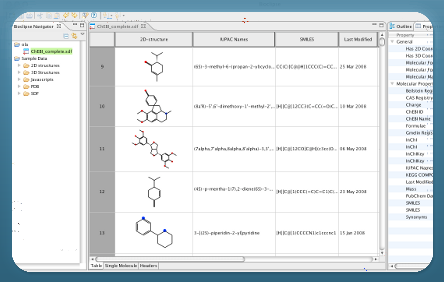
About Bioclipse
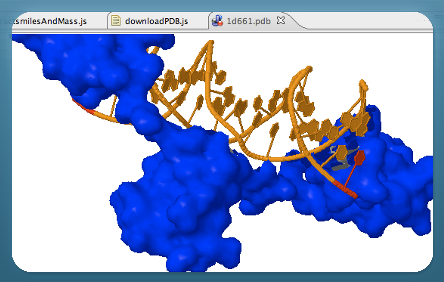
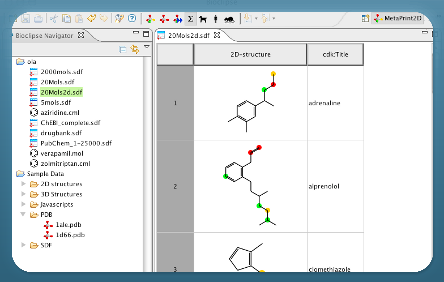
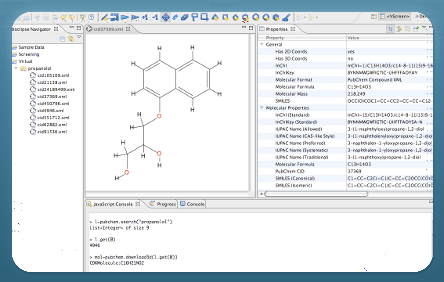
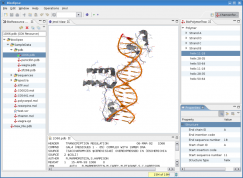 Bioclipse is a free and open source workbench for the life sciences.
Bioclipse is a free and open source workbench for the life sciences.
Bioclipse is based on the Eclipse Rich Client Platform (RCP) which means that Bioclipse inherits a state-of-the-art plugin architecture, functionality, and visual interfaces from Eclipse, such as help system, software updates, preferences, cross-platform deployment etc.